Does anyone have access to SPARC? They seem to have a very nice modeling software for Liquid-Liquid-Extraction (LLE).
Can anyone run logD simulations as seen here: http://www.archemcalc.com/video/logD.htm ?
Specific request (4 combos):
1) Substance psilocybin
2) Solvents DCM, Xylene, Hexane, limonene
3) Salt ammonium sulfate, salt
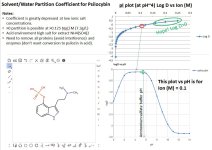
At the pI (~4), What is the expected distribution coefficient logD? Are there better solvent options out there that give a larger logD? Repeat calculations for psilocin at its isoelectric point (~9). As a bonus check Harmalol too.
Also for:
1) Substance psilocybin
2) Solvent acetone
3) Salt ammonium sulfate
What does the logD vs pH plot look like? Does the psilocybin tend to stay in the water layer overall?
Below are notes from using the free Marvin software. Unfortunately, one cannot change the solvent (standard octanol is used) nor the salt (NaCl) and the salt concentration does not go above 0.25M. The free Marvin software does not plot logD vs pH, but the plot can be generated by writing down the numbers it gives as the salt concentration is changed in the logP calculation under the "tools" menu.
Since the fee Marvin software is limited, any SPARC calculations would be appreciated. Thanks!
Can anyone run logD simulations as seen here: http://www.archemcalc.com/video/logD.htm ?
Specific request (4 combos):
1) Substance psilocybin
2) Solvents DCM, Xylene, Hexane, limonene
3) Salt ammonium sulfate, salt
At the pI (~4), What is the expected distribution coefficient logD? Are there better solvent options out there that give a larger logD? Repeat calculations for psilocin at its isoelectric point (~9). As a bonus check Harmalol too.
Also for:
1) Substance psilocybin
2) Solvent acetone
3) Salt ammonium sulfate
What does the logD vs pH plot look like? Does the psilocybin tend to stay in the water layer overall?
Below are notes from using the free Marvin software. Unfortunately, one cannot change the solvent (standard octanol is used) nor the salt (NaCl) and the salt concentration does not go above 0.25M. The free Marvin software does not plot logD vs pH, but the plot can be generated by writing down the numbers it gives as the salt concentration is changed in the logP calculation under the "tools" menu.
Since the fee Marvin software is limited, any SPARC calculations would be appreciated. Thanks!

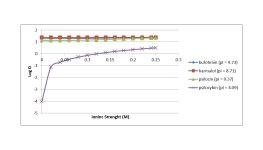
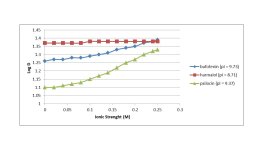
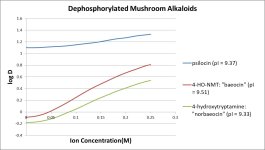
 shows the steepest slope on the graph. Does this relate to the internal hydrogen bonding mentioned in the
shows the steepest slope on the graph. Does this relate to the internal hydrogen bonding mentioned in the 
