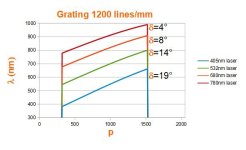
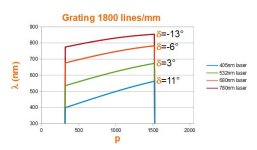
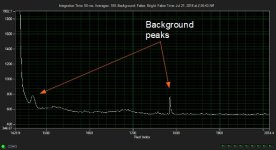
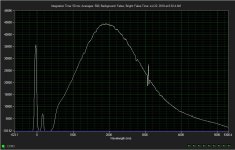
# OpticalRayTracer 9.6
#
* OpticalRayTracer Home Page
# 2018.07.18 11:16:00 EDT
program {
antialias = true
askBeforeDeleting = true
beamAngle = 0.000000e+00
beamCount = 5
beamWidth = 1
clipboardGraphicXSize = 1280
colorArrow = -2147483393
colorBaseline = -16760832
colorBeam = -4194304
colorGrid = 1086374080
colorHighBackground = -1
colorLensOutline = 276865279
colorLensSelected = 268484608
colorLightSource = -16776961
colorLowBackground = -16777216
colorTerminator = -16777216
decimalPlaces = 4
defaultWindowHeight = 600
defaultWindowWidth = 900
dispScale = 0.042214
dispersionBeams = 8
divergingSource = false
helpScrollPos = 0
interLensEpsilon = 1.000000e-06
intersectionArrowSize = 0.050000
inverse = false
maxIntersections = 64
selectedComponent = 6
selectedTab = 0
showControls = true
showGrid = true
snapValue = 0.000000e+00
surfEpsilon = 5.000000e-04
tableLineLimit = 500
virtualSpaceSize = 100.000000
windowX = 100
windowY = 100
xBeamRotationPlane = 0.000000e+00
xBeamSourceRefPlane = -30.000000
xOffset = -3.094207
yEndBeamPos = 1.800000
yOffset = -44.420187
yStartBeamPos = -1.800000
}
object {
active = true
angle = 0.000000e+00
centerThickness = 0.100000
dispersion = 59.000000
function = 2
ior = 1.520000
leftCurvature = 3
leftSphereRadius = 10.000000
leftZValue = 20.000000
lensRadius = 9.600000
name = Terminal Plane
rightCurvature = 3
rightSphereRadius = 10.000000
rightZValue = 20.000000
symmetrical = true
thickness = 0.100000
xPos = -19.997900
yPos = -46.114000
}
object {
active = true
angle = -42.500000
centerThickness = 0.100000
dispersion = 59.000000
function = 1
ior = 1.520000
leftCurvature = 3
leftSphereRadius = 6.000000
leftZValue = 20.000000
lensRadius = 4.000000
name = Mirror 1
rightCurvature = 3
rightSphereRadius = 6.000000
rightZValue = 20.000000
symmetrical = true
thickness = 0.100000
xPos = 0.000000e+00
yPos = 0.000000e+00
}
object {
active = true
angle = 95.000000
centerThickness = 0.552385
dispersion = 0.000000e+00
function = 0
ior = 1.520000
leftCurvature = 2
leftSphereRadius = 23.600000
leftZValue = 850.000000
lensRadius = 3.600000
name = Lens 1
rightCurvature = 2
rightSphereRadius = 23.600000
rightZValue = 850.000000
symmetrical = true
thickness = 0.000000e+00
xPos = -1.500000
yPos = -12.500000
}
object {
active = true
angle = 88.000000
centerThickness = 0.400000
dispersion = 59.000000
function = 1
ior = 1.520000
leftCurvature = 1
leftSphereRadius = 36.000000
leftZValue = 20.000000
lensRadius = 1.700000
name = Lens 2
rightCurvature = 1
rightSphereRadius = -36.000000
rightZValue = 20.000000
symmetrical = false
thickness = 0.400000
xPos = -5.260000
yPos = -52.500000
}
object {
active = true
angle = 84.000000
centerThickness = 1.600000
dispersion = -1.330000
function = 0
ior = 1.320000
leftCurvature = 3
leftSphereRadius = -20.700000
leftZValue = 0.000000e+00
lensRadius = 3.100000
name = Lens 3
rightCurvature = 3
rightSphereRadius = 6.000000
rightZValue = 20.000000
symmetrical = false
thickness = 1.600000
xPos = -8.195500
yPos = -36.658600
}
object {
active = true
angle = 67.000000
centerThickness = 0.100000
dispersion = 68.000000
function = 1
ior = 1.520000
leftCurvature = 3
leftSphereRadius = 6.000000
leftZValue = 20.000000
lensRadius = 1.900000
name = Lens 3
rightCurvature = 3
rightSphereRadius = 6.000000
rightZValue = 20.000000
symmetrical = false
thickness = 0.100000
xPos = -8.226700
yPos = -36.605100
}
object {
active = true
angle = 21.000000
centerThickness = 0.459292
dispersion = 59.000000
function = 1
ior = 1.520000
leftCurvature = 1
leftSphereRadius = 37.700000
leftZValue = 20.000000
lensRadius = 5.000000
name = Lens 2
rightCurvature = 1
rightSphereRadius = -45.800000
rightZValue = 20.000000
symmetrical = false
thickness = 0.400000
xPos = 1.982600
yPos = -46.598500
}